Note
Go to the end to download the full example code.
Working with netCDF groups#
This is an example about how to work with netCDF groups. Xarray does not natively read netCDF group files, but it does have the ability to read the data with a few independent calls.
Author: Ken Kehoe
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
from arm_test_data import DATASETS
from act.io.arm import read_arm_netcdf
from act.plotting import TimeSeriesDisplay
# This data file is a bit complicated with the group organization. Each group
# will need to be treated as a different netCDF file for reading. We can read each group
# independently and merge into a single Dataset to use with standard Xarray or ACT methods.
# Top Level:
# data
#
# Groups:
# - data
# - light_absorption
# - instrument
# - particle_concentration
# - instrument
# - light_scattering
# - instrument
filename = DATASETS.fetch(
'ESRL-GMD-AEROSOL_v1.0_HOUR_MLO_s20200101T000000Z_e20210101T000000Z_c20210214T053835Z.nc'
)
# We start by reading the location information from the top level of the netCDF file.
# This is a standard Xarary call without a group keyword. Only the top level data is read.
# The returned Dataset will contain all top level variables and top level global attributes.
# We will use the direct Xarray call to read data since there is no time dimension at the top
# level so we don't need ACT to do anything for us.
ds_location = xr.open_mfdataset(filename)
# Second, we read the 'data' group. This has a time dimension so we can use ACT to manage
# correctly reading and formatting that information. We need to specify the 'data' group
# to be read. This will read the data group but not the sub-group.
ds_data = read_arm_netcdf(filename, group='data')
# Third, we read the 'light_scattering' sub-group. We can read sub-groups
# by using standard Linux directory path notation. Notice the light_scattering group is a
# sub-group to 'data'.
ds_ls = xr.open_mfdataset(filename, group='data/light_scattering')
# We can also read the third group 'instrument'. This only contains a small amount of information
# about the instrument used to collect the data for 'light_scattering' and has no
# dimensions, only scalar values.
ds = xr.open_mfdataset(filename, group='data/light_scattering/instrument')
# Since the dimensionality aligns we can merge the three Datasets into a single Dataset.
ds = xr.merge([ds_data, ds_ls, ds_location])
# Since the data file contains data for a full year we can subset the Dataset to be easier
# to work with and process faster.
ds = ds.sel(time=slice('2020-01-01T00:00:00', '2020-01-04T23:59:59'))
# The data is written with dimensionality in reverse order from what ACT expects. We need to
# reverse the dimensionality order.
ds = ds.transpose()
# Since we want to only plot the data from one of the cut sizes we can subset the Dataset
# to only have data where the cut_size variable is set to a single value. This will reduce
# the dimensionality from 3 to 2 or 2 to 1 dimensions on the variables. Take note that
# the value of cut size is not the um value, it is the index. Since the index values
# are 0 or 1, 0 = 1 um and 1 = 10 um. We are subsetting for less than 10 um.
ds = ds.isel(cut_size=1)
# netCDF has a default _FillValue that is used to indicate when the values are missing. There
# is a known issue with float precision that does not correctly set the _FillValue to NaN
# when reading. We need to set the obviously incorrect values to NaN for the data variables
# that can have this problem.
for var_name in ds.data_vars:
try:
data = ds[var_name].values
data[ds[var_name].values >= 9e36] = np.nan
ds[var_name].values = data
except np._core._exceptions._UFuncNoLoopError:
pass
# This is a querk of reading the data. If we want to plot the day/night background correctly
# we need to delete these global attribute.
del ds.attrs['_file_dates']
del ds.attrs['_file_times']
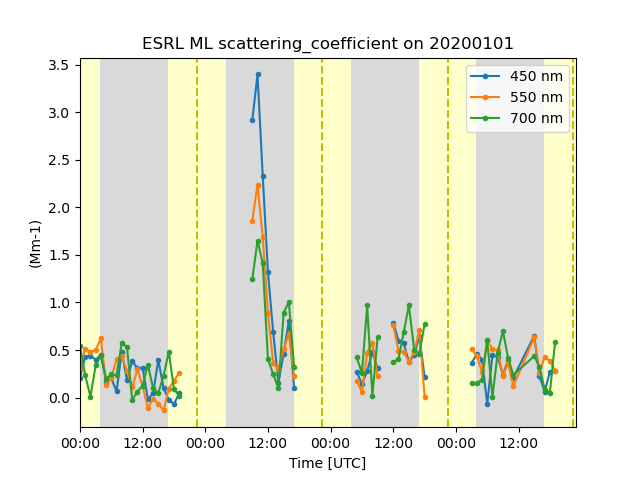
# Since the data variable has a second dimension of wavelength and we want to plot each one as a
# line plot, we will pass force_line_plot=True to force the second dimension to be removed from
# the time series plot. We will want to set the labels to view on the plot by extracting the values
# from the wavelenght dimension and pass into the plotting call.
labels = [f"{int(wl)} {ds['wavelength'].attrs['units']}" for wl in ds['wavelength'].values]
display = TimeSeriesDisplay({'ESRL ML': ds})
display.plot(
'scattering_coefficient', day_night_background=True, force_line_plot=True, labels=labels
)
plt.show()
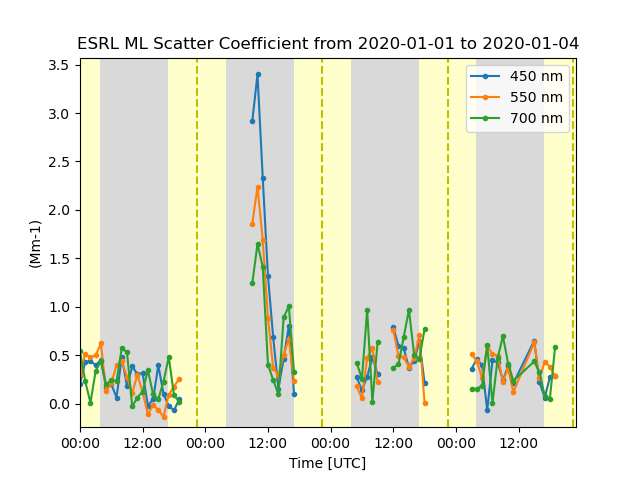
# A second option is to extract the wavelength dimension form the variable and create a new variable
# to plot. We are selecting the wavelength by using index so a value of 1 = 550 um. We need to
# provide a new variable name not already in use and correctly describes the data.
ds['scattering_coefficient_450'] = ds['scattering_coefficient'].isel(indexers={'wavelength': 0})
ds['scattering_coefficient_550'] = ds['scattering_coefficient'].isel(indexers={'wavelength': 1})
ds['scattering_coefficient_700'] = ds['scattering_coefficient'].isel(indexers={'wavelength': 2})
display = TimeSeriesDisplay({'ESRL ML': ds})
title = 'ESRL ML Scatter Coefficient from 2020-01-01 to 2020-01-04'
display.plot('scattering_coefficient_450', label=labels[0], day_night_background=True)
display.plot('scattering_coefficient_550', label=labels[1])
display.plot('scattering_coefficient_700', label=labels[2], set_title=title)
plt.show()
Total running time of the script: (0 minutes 1.575 seconds)