Note
Go to the end to download the full example code.
Plotting Baseline Surface Radiation Network (BSRN) QC Flags#
Simple example for applying BSRN QC and plotting the data and the corresponding QC flags using colorblind friendly colors. https://bsrn.awi.de/data/quality-checks/
Author: Ken Kehoe
from arm_test_data import DATASETS
from matplotlib import pyplot as plt
import act
# Read in data and convert from ARM QC standard to CF QC standard
filename_brs = DATASETS.fetch('sgpbrsC1.b1.20190705.000000.cdf')
ds = act.io.arm.read_arm_netcdf(filename_brs, cleanup_qc=True)
# Creat Plot Display and plot data including embedded QC from data file
variable = 'down_short_hemisp'
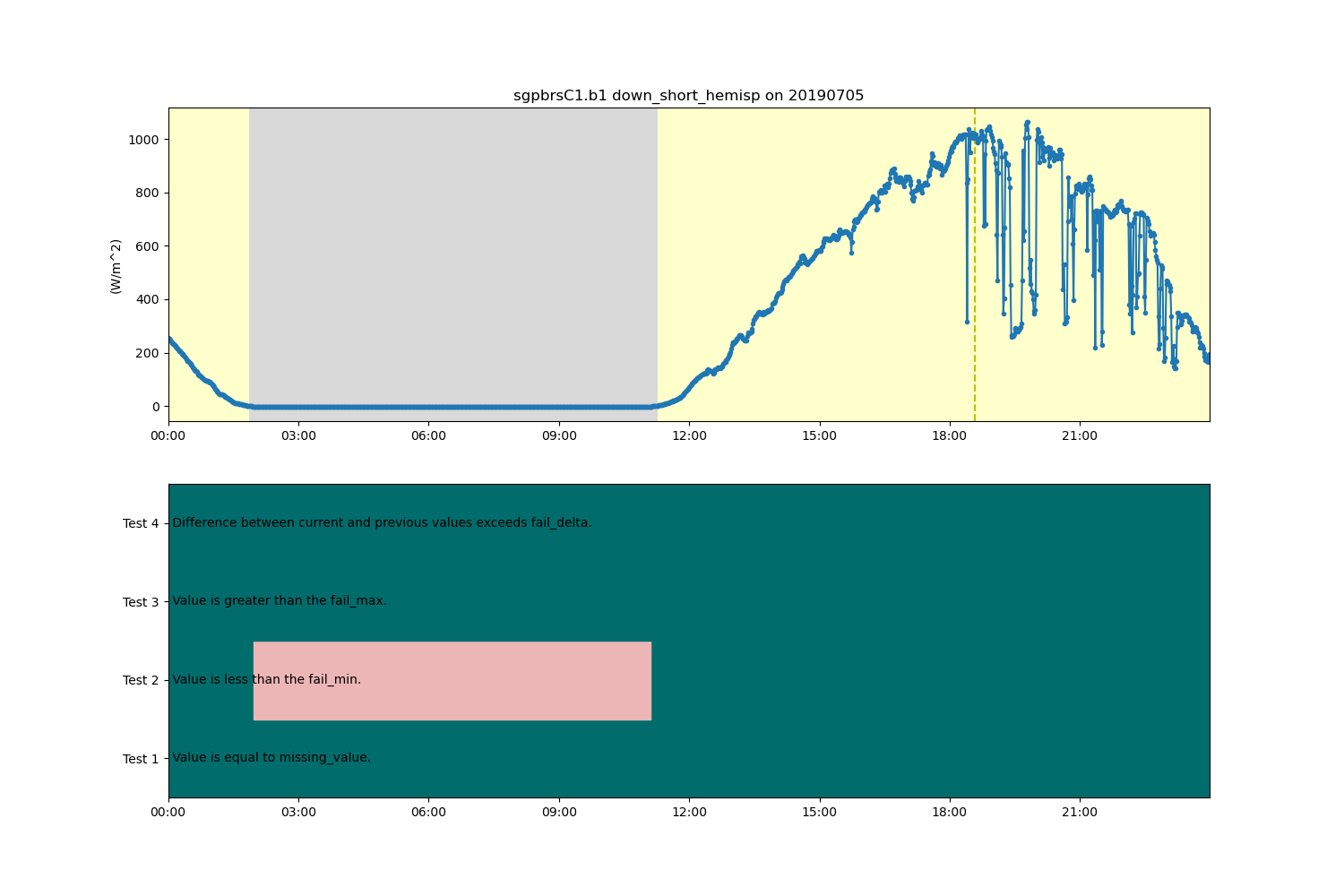
display = act.plotting.TimeSeriesDisplay(ds, figsize=(15, 10), subplot_shape=(2,))
# Plot radiation data in top plot
display.plot(variable, subplot_index=(0,), day_night_background=True, cvd_friendly=True)
# Plot ancillary QC data in bottom plot
display.qc_flag_block_plot(variable, subplot_index=(1,), cvd_friendly=True)
plt.show()
# Add initial BSRN QC tests to ancillary QC varialbles. Use defualts for
# test set to Physicall Possible and use_dask.
ds.qcfilter.bsrn_limits_test(
gbl_SW_dn_name='down_short_hemisp',
glb_diffuse_SW_dn_name='down_short_diffuse_hemisp',
direct_normal_SW_dn_name='short_direct_normal',
glb_SW_up_name='up_short_hemisp',
glb_LW_dn_name='down_long_hemisp_shaded',
glb_LW_up_name='up_long_hemisp',
)
# Add initial BSRN QC tests to ancillary QC varialbles. Use defualts for
# test set to Extremely Rare" and to use Dask processing.
ds.qcfilter.bsrn_limits_test(
test='Extremely Rare',
gbl_SW_dn_name='down_short_hemisp',
glb_diffuse_SW_dn_name='down_short_diffuse_hemisp',
direct_normal_SW_dn_name='short_direct_normal',
glb_SW_up_name='up_short_hemisp',
glb_LW_dn_name='down_long_hemisp_shaded',
glb_LW_up_name='up_long_hemisp',
use_dask=True,
)
# Add comparison BSRN QC tests to ancillary QC varialbles. Request two of the possible
# comparison tests.
ds.qcfilter.bsrn_comparison_tests(
['Global over Sum SW Ratio', 'Diffuse Ratio'],
gbl_SW_dn_name='down_short_hemisp',
glb_diffuse_SW_dn_name='down_short_diffuse_hemisp',
direct_normal_SW_dn_name='short_direct_normal',
)
# Add K-tests QC to ancillary QC variables.
ds.qcfilter.normalized_rradiance_test(
[
'Clearness index',
'Upper total transmittance',
'Upper direct transmittance',
'Upper diffuse transmittance',
],
dni='short_direct_normal',
dhi='down_short_hemisp',
ghi='down_short_diffuse_hemisp',
)
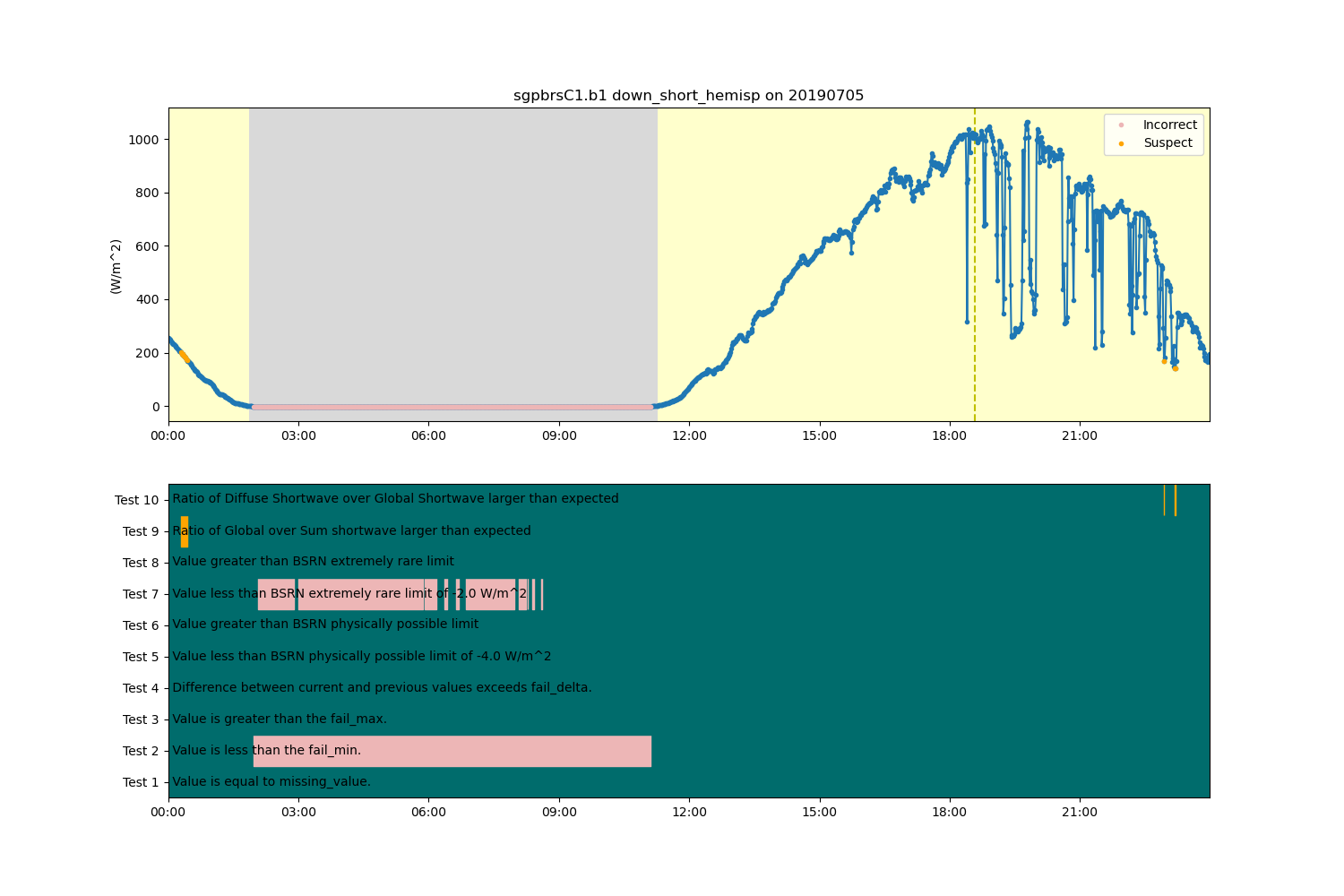
# Creat Plot Display and plot data including embedded QC from data file
variable = 'down_short_hemisp'
display = act.plotting.TimeSeriesDisplay(ds, figsize=(15, 10), subplot_shape=(2,))
# Plot radiation data in top plot. Add QC information to top plot.
display.plot(
variable,
subplot_index=(0,),
day_night_background=True,
assessment_overplot=True,
cvd_friendly=True,
)
# Plot ancillary QC data in bottom plot
display.qc_flag_block_plot(variable, subplot_index=(1,), cvd_friendly=True)
plt.show()
Total running time of the script: (0 minutes 1.414 seconds)